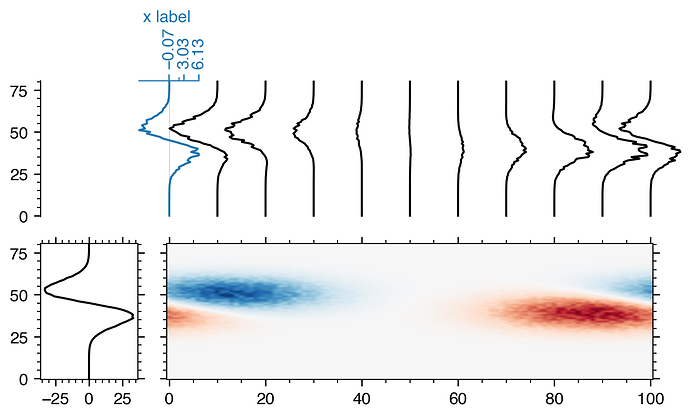
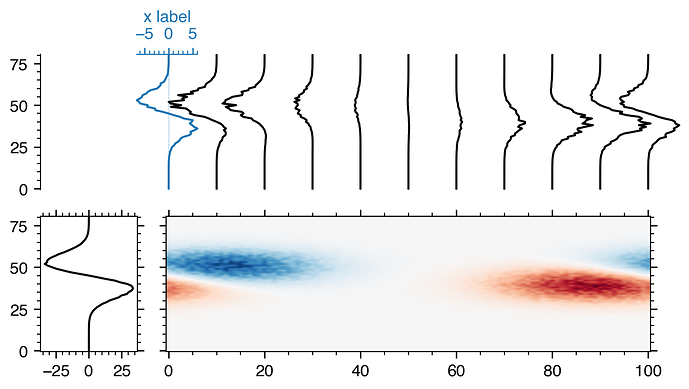
Thanks a lot @jklymak!
Adding an individual axes with blended transform is something I would have never thought of myself.
Adding an axes for every profile does make plotting a bit slow though. I went with adding an axes for only the first profile and plotting the others in the parent axes. ~1 s vs. ~70 ms on a MacBook Pro, 15-inch, 2016, 2.7 GHz Intel Core i7.
Also, I am not sure what is the advantage of using an inset axes. I guess the same would work with fig.add_axes.
I am posting the full code for the more complex example in case someone (probably future-me) runs into this problem again:
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.transforms import blended_transform_factory
# make some sample data
dx = dy = 1
y = np.arange(80, 0 - dy, -dy)
x = np.arange(0, 100 + dx, dx)
x_offsets = np.linspace(0, 100, 11)
xx, yy = np.meshgrid(0.05 * (x + 10), 0.1 * (y - 40))
data1 = np.exp(-xx**2 - yy**2) - np.exp(-(xx - 1)**2 - (yy - 1)**2)
xx, yy = np.meshgrid(0.05 * (x - 90), 0.1 * (y - 40))
data2 = np.exp(-xx**2 - yy**2) - np.exp(-(xx - 1)**2 - (yy - 1)**2)
data = data1 + data2
data += np.random.rand(data.shape[0], data.shape[1]) * 0.5 * data
extent = (x[0] - 0.5 * dx, x[-1] + 0.5 * dx, y[-1] - 0.5 * dy, y[0] + 0.5 * dy)
# set up the plot
fig, ax = plt.subplots(
2, 2, sharey=True, figsize=(8, 4),
gridspec_kw=dict(width_ratios=(0.2, 1), wspace=0.1)
)
axTL = ax[0, 0]
axTR = ax[0, 1]
axBL = ax[1, 0]
axBR = ax[1, 1]
trans = blended_transform_factory(axTR.transData, axTR.transAxes)
data_abs_max = np.abs(data).max()
im = axBR.imshow(data, 'RdBu_r', vmin=-data_abs_max, vmax=data_abs_max,
extent=extent, aspect='auto', interpolation='bilinear')
axBR.axis(extent)
axBL.plot(data.sum(axis=1), y, 'k')
scale = 8
for offset in x_offsets:
profile = data[:, int(offset / dx)]
profile = scale * profile
xmin, xmax = profile.min(), profile.max()
if offset == 0:
bounds = (offset + xmin, 0, xmax - xmin, 1)
inset_ax = axTR.inset_axes(bounds, transform=trans)
inset_ax.set_ylim(axTR.get_ylim())
inset_ax.set_xlim(xmin, xmax)
color = 'tab:blue'
inset_ax.axvline(0, color=color, ls='dotted', lw=0.5)
inset_ax.plot(profile, y, color, clip_on=False, zorder=1)
inset_ax.set_facecolor('none')
inset_ax.spines['left'].set_visible(False)
inset_ax.spines['bottom'].set_visible(False)
inset_ax.spines['right'].set_visible(False)
inset_ax.spines['top'].set_color('tab:blue')
inset_ax.tick_params(
'both', which='both',
top=True, left=False, right=False, bottom=False,
labeltop=True, labelleft=False,
color='tab:blue', labelcolor='tab:blue'
)
inset_ax.set_xlabel('x label', color='tab:blue')
inset_ax.xaxis.set_label_position('top')
inset_ax.xaxis.tick_top()
else:
color = 'k'
axTR.plot(profile + offset, y, color, clip_on=False, zorder=0)
# remove unwanted spines and ticks
axTR.axis('off')
axTL.spines['top'].set_visible(False)
axTL.spines['right'].set_visible(False)
axTL.spines['bottom'].set_visible(False)
axTL.tick_params('both', which='both', top=False, right=False, bottom=False,
labelbottom=False)
axBR.tick_params('both', which='both', labelleft=False)
axTR.axis(extent)